Single Cell Transcriptome Analysis (10xgenomics)
Table of Contents
- Overview
- Cellranger software and versions
- Reference Genome
- Information about cellranger run configuration
- Alignment summary metrics
- Singlecell QC check using Scanpy
- UCSC Cell Browser
- Software and versions
- List of resources
- Change logs
Overview
Cellranger pipeline from 10Xgenomics is used for running primary analysis for the single cell transcriptome samples (currently, only the 3' single cell RNA-Seq data is supported). A list of the output files from this pipeline can be found here.
Cellranger software and versions
Cellranger command line
Example cellranger count command.
cellranger count
--fastqs=/fastq_directory_path
--id=ID
--transcriptome=/path/transcriptome
--nopreflight
--disable-ui
--localcores=14
--localmem=64
Reference Genome
Following reference genomes are used for running Cellranger data analysis pipeline:
-
Human: A custom reference genome using the hg38 genome build and Gencode (v30) gene sets
-
Human (pre-mRNA): A pre-mRNA reference genome using the hg38 genome build and Gencode (v30) gene sets
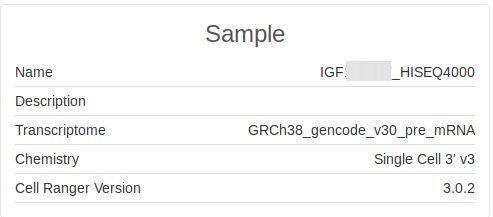
Information about cellranger run configuration
Check the Sample information section of the Cellranger html report for more information regarding the reference genome build, Single cell chemistry version and Cellranger version information.
Alignment summary metrics
A multiqc report for the alignment bam is produced (per sample) combining the following Picard and Samtools metrics.
- Picard CollectAlignmentSummaryMetrics
- Picard CollectBaseDistributionByCycle
- Picard CollectGcBiasMetrics
- Picard QualityScoreDistribution
- Picard CollectRnaSeqMetrics
- Samtools stats
- Samtools idxstats
Singlecell QC check using Scanpy
We use Scanpy to generate per sample QC report for the single cell data following this tutorial: Clustering 3K PBMCs. We have implemented a Jupyter notebook based QC report which can be run within a Docker or Singularity container. We execute this notebook based implementation for each of the single cell samples and store a html version of the report with all the codes and plots. These notebook resources can be accessed from the ‘Analysis’ tab of the ‘IGF QC Report’ page.
Please feel free to check our implementation of the QC report on Github scanpy-notebook-image for further detail or to try out the example notebooks on binder. A list of all our notebook based resources can be found this this page: Notebook resources.
UCSC Cell Browser
We have implemented an offline version of UCSC Cell Browser and generate the server resources for individual samples. Similar to the Scanpy report pagess, these browser directories can be accessed from the ‘Analysis’ tab of the ‘IGF QC Report’ page.
Software and versions
List of resources
- Custom reference genome building
- Reference genome for hg19 and mm10
- Cellranger output files
- Scanpy
- UCSC Cell Browser
- Github: scanpy-notebook-image
- Notebook resources
Change logs
- 3rd Feb 2020
- Moved to notebook based Scanpy QC report
- Updated Scanpy to v1.4 to v1.4.5
- Added UCSC Cell Browser
- 25th June 2019
- Added 3D UMAP plot
- Added pre-mRNA reference transcriptome option for Cellranger
- Moved to Gencode v30 build from v28
- 24th April 2019
- Changed Cellranger version from 2.2.0 to 3.0.2
- Changed Scanpy version from 1.2.2 to 1.4
- Moved Scanpy command from help page and added them to the reports
- 24th August 2018
- Added Scanpy report